Learn More about Tuffy
Users' Manual VLDB 2011 Paper Slides Technical Report
Installation Guide
Tuffy relies on Java and PostgreSQL. Since they are available on most commonly used platforms, you can run Tuffy on Linux, Windows, or Mac OS. This page guides you on how to install and configure those prerequisites as well as Tuffy itself.
Install Java Runtime
If you don't have JRE 1.6 (or higher) on your machine, please go to the Java website to download and install it.
Install and Configure PostgreSQL
-
If you don't have PostgreSQL 8.4 (or higher) on your machine, please
download and install it. Let PG_DIST be the path where you unpacked the source distribution. Let PG_PATH be the location where you want to install
PostgreSQL at. Run the following commands:
cd PG_DIST ./configure --prefix=PG_PATH gmake; gmake install cd PG_PATH/bin initdb -D PG_PATH/data postmaster -D PG_PATH/data & createdb test psql test
initdbinitializes a directory to store databases;postmasterlaunches the PostgreSQL server daemon;createdbcreates a new database (with name 'test' in our example); andpsqltakes you to the interactive console of PostgreSQL, where you can issue numerous kinds of SQL queries. Type '\h' for help, and '\q' to quit. -
Install additional modules; we only need 'intarray' and 'intagg':
cd PG_DIST/contrib/intarray gmake; gmake install cd PG_DIST/contrib/intagg gmake; gmake install
-
Create a PostgreSQL super user with name, say, "tuffer". Look here, or simply run the following command:
PG_PATH/bin/createuser -s -P tuffer
You will be promoted for a password. Henceforth let's assume the password is "strongPasswoRd". -
Create a database with name, say, "tuffydb". Look here, or simply run the following command:
PG_PATH/bin/createdb tuffydb PG_PATH/bin/createlang plpgsql tuffydb
Download and Configure Tuffy
- Grab Tuffy from the download page.
-
After unpacking, you should see a configuration file named "tuffy.conf" (see the figure below; lines beginning with '#' are comments). The four parameters in it are all you need to configure Tuffy. They control the PostgreSQL connection and the (temporary) working directory to be used by Tuffy. Tuffy will write temporary data to both places at runtime and clean them up when finishing. Make sure that they are going to be used by Tuffy exclusively. The default values are as follows.
# JDBC connection string; must be PostgreSQL db_url = jdbc:postgresql://localhost:5432/tuffydb # Database username; must be a superuser db_username = tuffer # The password for db_username db_password = strongPasswoRd # The working directory; Tuffy may write sizable temporary data here dir_working = /tmp/tuffy-workspace
- At this point, you should be good to go and be able to run Tuffy smoothly. However, if Tuffy complains that it cannot connect to PostgreSQL, it might be that the database was rejecting connections based on the IP of the machine on which you run Tuffy. Fixes can be found, e.g., here.
Build Tuffy
Instead of downloading the binaries, you may also download the Tuffy sources, and build the jar using the following instructions:ant ant dist
The first ant builds java classes in, while the second ant packages the jar.
Usage Guide
We use a simple example to illustrate how to use Tuffy. The input/output formats as well as command options are compatible with Alchemy.
Input
Conceptually, the input consists of three parts: the MLN program, the evidence, and the query (as shown above). The program and the evidence are each specified in one or more plain text files. The query may be specified with text files and/or the command line.

The input format of Tuffy is compatible with that of Alchemy. For example, a program file consists of predicate definitions and weighted clauses. Terms that begin with a lower-case letter are variables, while those beginning with an upper-case letter are constants. Existential quantifiers are supported. We do introduce a syntactic sugar, though: A predicate definition preceded by "*" is considered to have the closed world assumption (CWA); i.e., all ground atoms of this predicate not listed in the evidence are false. In the sample program above, we make such an assumption with the predicate Friends. Alternatively, one may also specify CWA predicates with the "-cw" option in the command line.
An evidence file is a list of ground atoms. Each atom is deemed to be true unless it's preceded by "!". In this particular example, the line "!Friends(Gary, Frank)" is superfluous since the predicate Friends is closed.
Suppose that our query is to determine the set of people who have cancer, which can be specified with the atom Cancer(x). We can specify this query in either of the following two ways. First, we may use the command line option -q Cancer(x) or -q Cancer. Second, we may write a text file with the content Cancer(x) and specify -queryFile fileName in the command line, where fileName is the path to the text file. Sometimes the query is more complex than just a couple of atoms; in this case, the second approach can be convenient.
Inference
There are two types of inference with MLNs: MAP inference and marginal inference. By default, Tuffy runs MAP inference to produce a most likely world; but you can use "-marginal" to instruct Tuffy to run marginal inference instead. More concretely, suppose you want to find the most likely set of people with cancer -- which is MAP inference -- then run the following command:
java -jar tuffy.jar -i samples/smoke/prog.mln -e samples/smoke/evidence.db -queryFile samples/smoke/query.db -r out.txt
which would produce terminal output similar to the screenshot below:
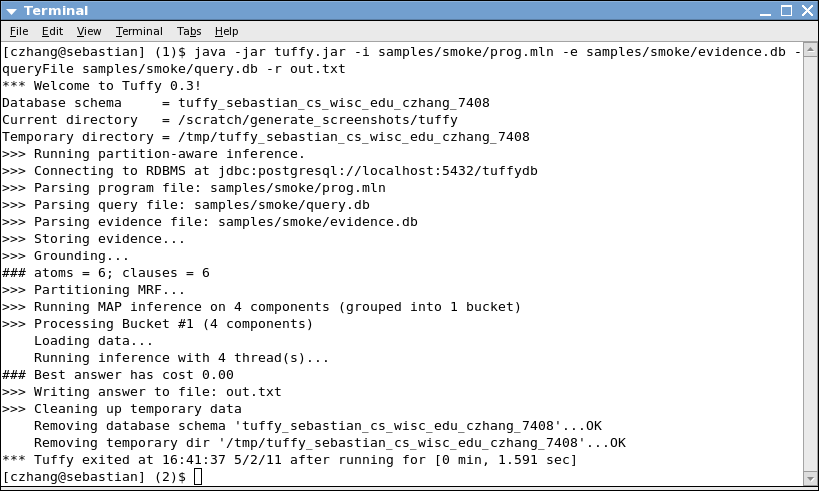
We have divided the screenshot into five segments to summarize the steps involved in
this invocation of Tuffy: Setup, Parsing, Grounding, Inference, and Clean-up.
To better understand the algorithms involved in Grounding and Inference, please
read the manual and/or refer to our technical report.
The inference result is written to a file named out.txt, whose content should resemble the text below:
Cancer(Anna) Cancer(Bob) Cancer(Edward) Cancer(Frank)
This world actually has a cost 0; i.e., all rules are fully satisfied. For example, one can verify that every smoker has cancer, complying with the first rule in the MLN program. Note that the output is a projection of the MAP world on the query. For example, in this particular case, even though the inference algorithm has determined that Bob and Frank are also smokers, these facts are not shown in the output file. Had the query contained both Cancer(x) and Smokes(x), the output would also contain Smokes(Bob) and Smokes(Frank).
Alternatively, you may want to estimate the probability that each person has cancer -- which is marginal inference. In this case, run
java -jar tuffy.jar -marginal -i samples/smoke/prog.mln -e samples/smoke/evidence.db -queryFile samples/smoke/query.db -r out.txt
which would produce a results like
0.75 Cancer(Edward) 0.65 Cancer(Anna) 0.50 Cancer(Bob) 0.45 Cancer(Frank)
The numbers before each atom are the estimated marginal probabilities. Atoms absent from the output have probability 0.
Command Options
Here is a list of the command line options:
USAGE:
-activateAll : Mark all unknown atoms as active during grounding.
-conf VAL : Path of the configuration file. Default='./tuffy.conf'
-cw (-closedWorld) VAL : Specify closed-world predicates. Separate with comma.
-dMaxIter N : Max number of iterations for learning. DEFAULT=500
-db VAL : The database schema from which the evidence is loaded.
-dbNeedTranslate : Whether we can directly use tables in this DB
schema. See -db option.
-dontBreak : Forbid WalkSAT steps that would break hard
clauses. DEFAULT=FALSE
-dribble VAL : File where terminal output will be written to.
-dual : Run both MAP and marginal inference. Results will
be written to $out_file.map and $out_file.marginal
respectively.
-e (-evidence) VAL : REQUIRED. Input evidence file(s). Separate with comma.
-gz : Compress output files in gzip.
-help : Display command options.
-i (-mln) VAL : REQUIRED. Input MLN program(s). Separate with comma.
-keepData : Keep the data in the database upon exiting.
-learnwt : Run Tuffy in discriminative weight learning mode.
-lineHeader VAL : Precede each line printed on the console with this string.
-marginal : Run marginal inference with MC-SAT.
-maxFlips N : Max number of flips per try. Default=[#atoms] X 10
-maxTries N : Max number of tries in WalkSAT. Default=1
-mcsatParam N : Set x; each step of MC-SAT retains each
non-violated clause with probability 1-exp(-|weight|*x).
DEFAULT=1.
-mcsatSamples N : Number of samples used by MC-SAT. Default=20.
-minProb N : Mininum probability for output of marginal inference.
-nopart : Disable MRF partitioning. (Partitioning is enabled by default.)
-o (-r, -result) VAL : REQUIRED. Output file.
-printResultsAsPrologFacts : print Results As Prolog Facts.
-q (-query) VAL : Query atom(s). Separate with comma.
-queryFile VAL : Input query file(s). Separate with comma.
-threads N : The max num of threads to run in parallel.
Default = #available processors
-timeout N : Timeout in seconds.
-verbose N : Verbose level (0-3). Default=0
Tuffy in Depth
Browse the java doc online.
Check out the unit tests coverage report.
Read the technical report.
Download the user manual.
Other Resources
The learning component in the current version of Tuffy is rather basic. For users who want to transform pre-weight learning rules with Alchemy's '+' notation, there is a nice Python script written by Oscar Sjober that can do the job.
